GWAS-Based Identification of New Loci for Milk Yield, Fat, and Protein in Holstein Cattle
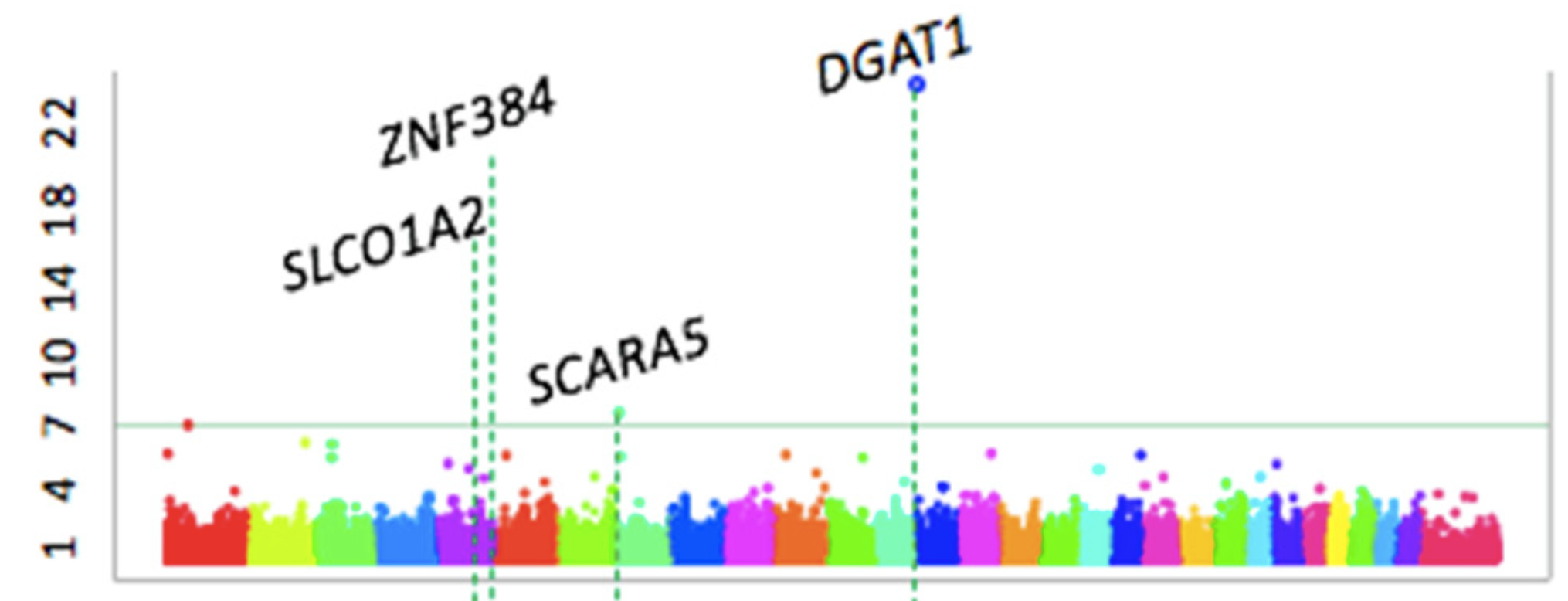
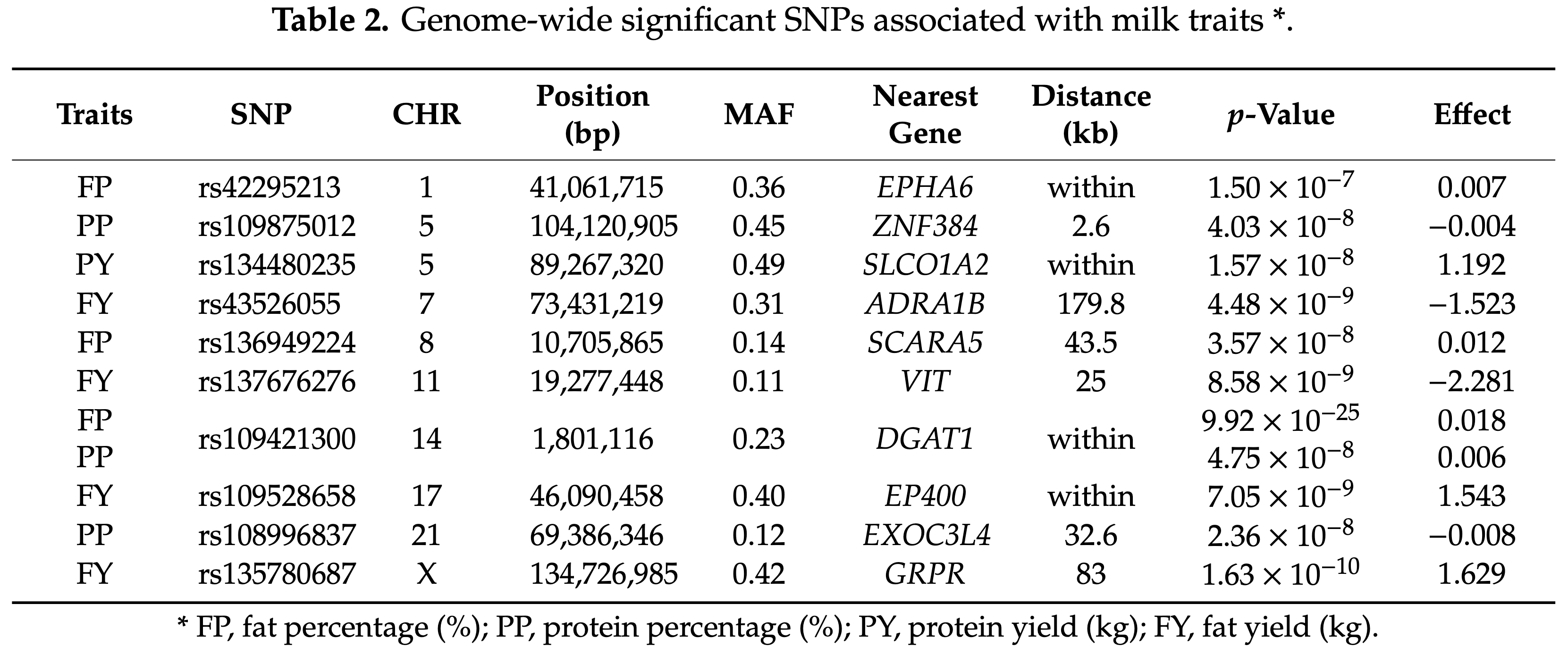
Identification of Significant Loci for Milk Quality
The study detected ten SNPs that surpassed the genome-wide significance threshold (p<4.0×10-7) for milk fat and protein traits, while no significant markers were found for milk yield. Among these, six SNPs were located within previously reported quantitative trait loci (QTL) regions, confirming the major effect of the DGAT1 gene on BTA 14 for both fat and protein percentages, while also identifying novel candidate genes such as EPHA6 (fat percentage) and SLCO1A2 (protein yield).
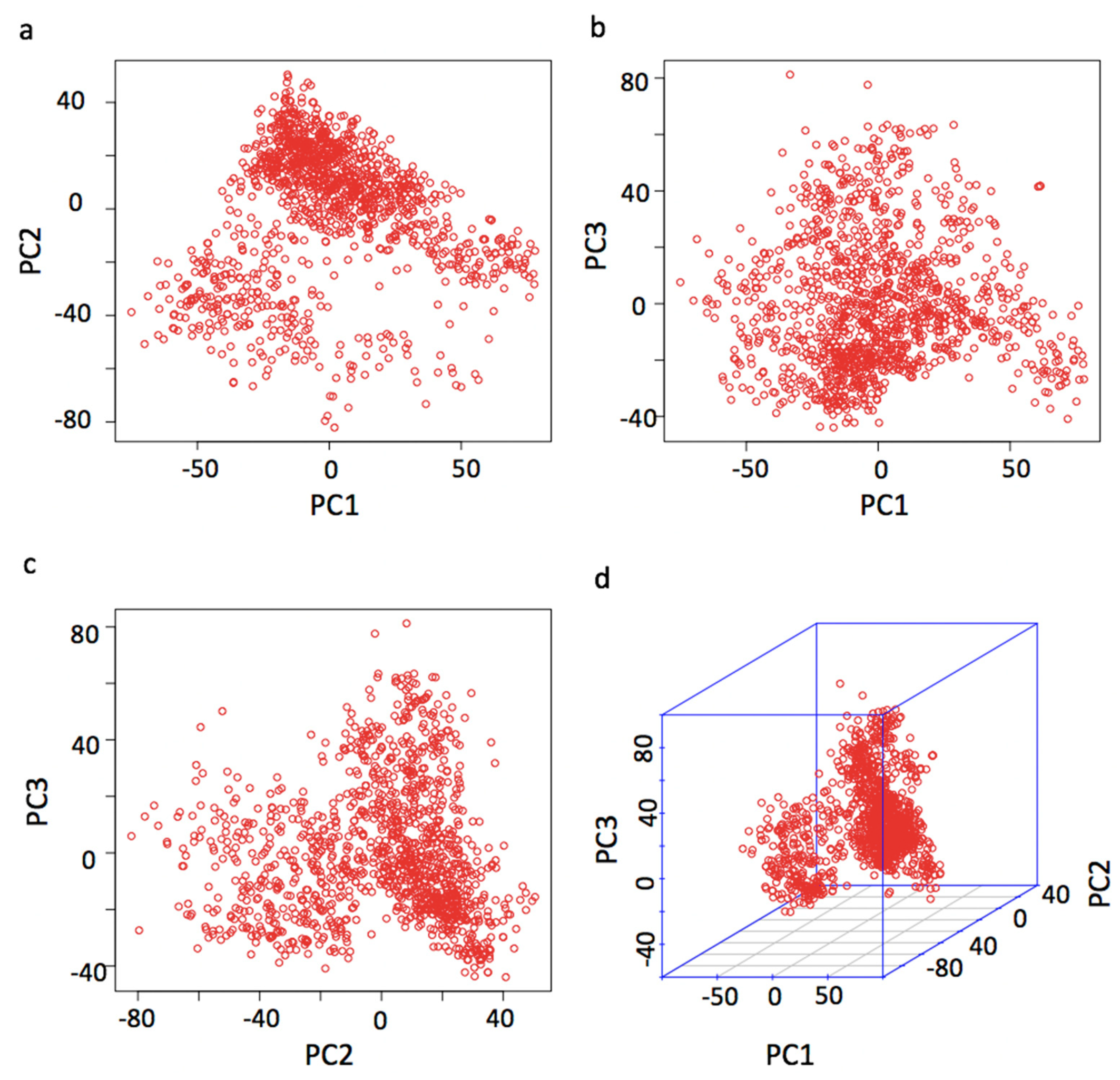
Controlling Population Stratification
Principal component analysis (PCA) revealed a stratified population structure within the herd, likely due to the use of imported semen and varying genetic backgrounds. To address this, the study employed the Fixed and random model Circulating Probability Unification (FarmCPU) method with the first three principal components fitted as covariates; this approach effectively controlled for false positives, as evidenced by the Quantile-Quantile (QQ) plots showing minimal inflation.
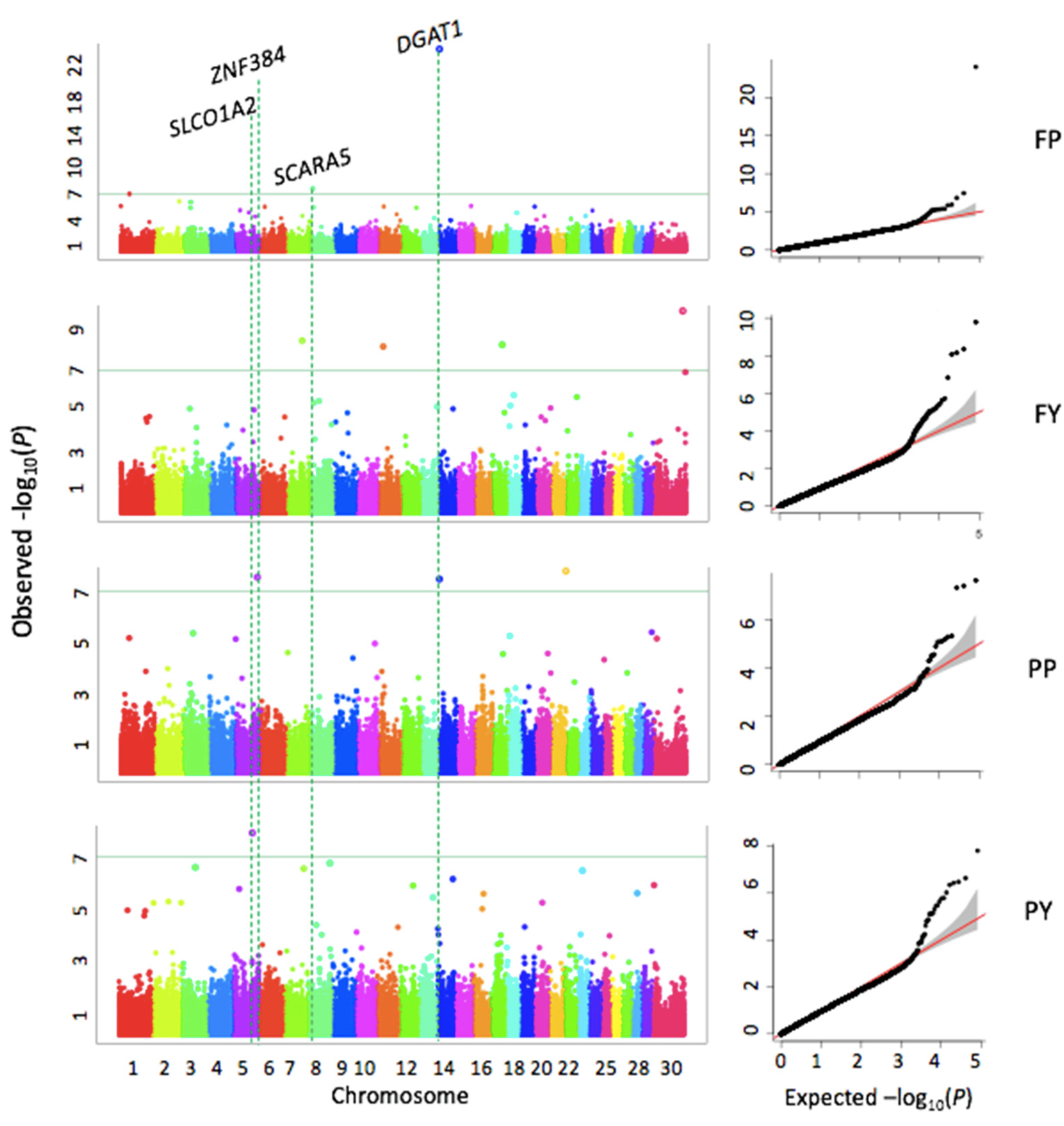
Pleiotropic Effects Across Yield Traits
The analysis revealed pleiotropic genetic effects where specific genomic regions influenced multiple traits simultaneously. For instance, in addition to the DGAT1 gene affecting both fat and protein percentages, a heatmap of p-values indicated overlapping significant regions for milk yield, protein yield, and fat yield on chromosomes 1, 2, 3, 11, 17, 20, and 22, suggesting shared genetic regulatory mechanisms for these correlated production traits.
