iPat: intelligent prediction and association tool for genomic research
Intuitive Graphical User Interface
To overcome the steep learning curve associated with Command-Line Interfaces (CLI), iPat implements a 'drag and drop' system where users simply use a pointing device to manage data files and project icons within a desktop-like frame. The interface transforms complex R-library parameters into accessible graphical elements—such as sliders and checkboxes—allowing users to easily configure input parameters, define models, and visualize results without writing code.
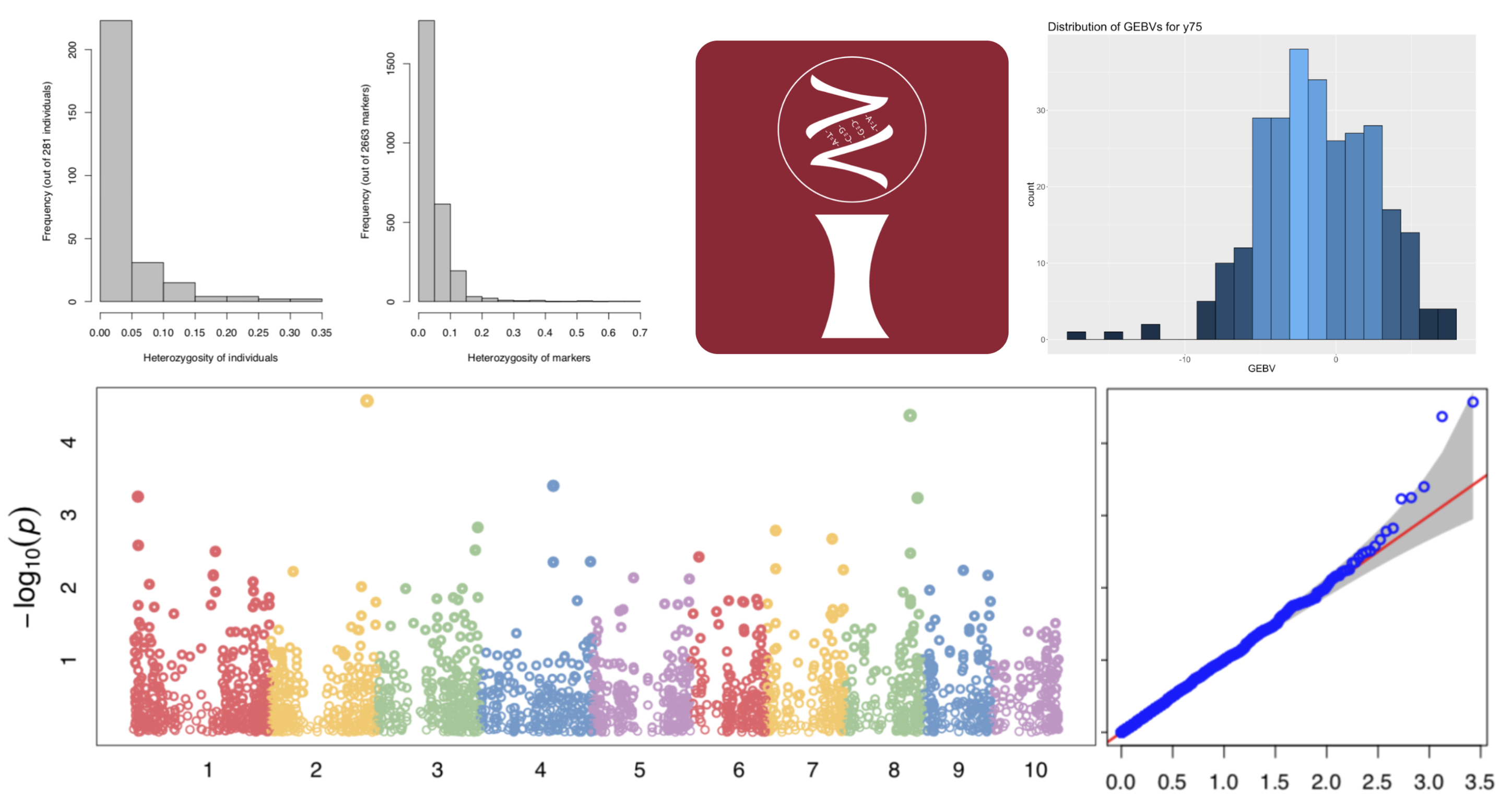
Integration of Robust R Libraries and Methods
iPat serves as an intelligent wrapper that seamlessly integrates multiple widely-used CLI packages, including R-based libraries like GAPIT, rrBLUP, and BGLR, as well as C-driven tools like PLINK and FarmCPU. This integration enables users to deploy advanced statistical methods for GWAS and genomic prediction (including GWAS-assisted prediction) within a unified environment, automatically translating user inputs into R scripts to ensure robust genomic analytic results and visualization of accuracy distributions.
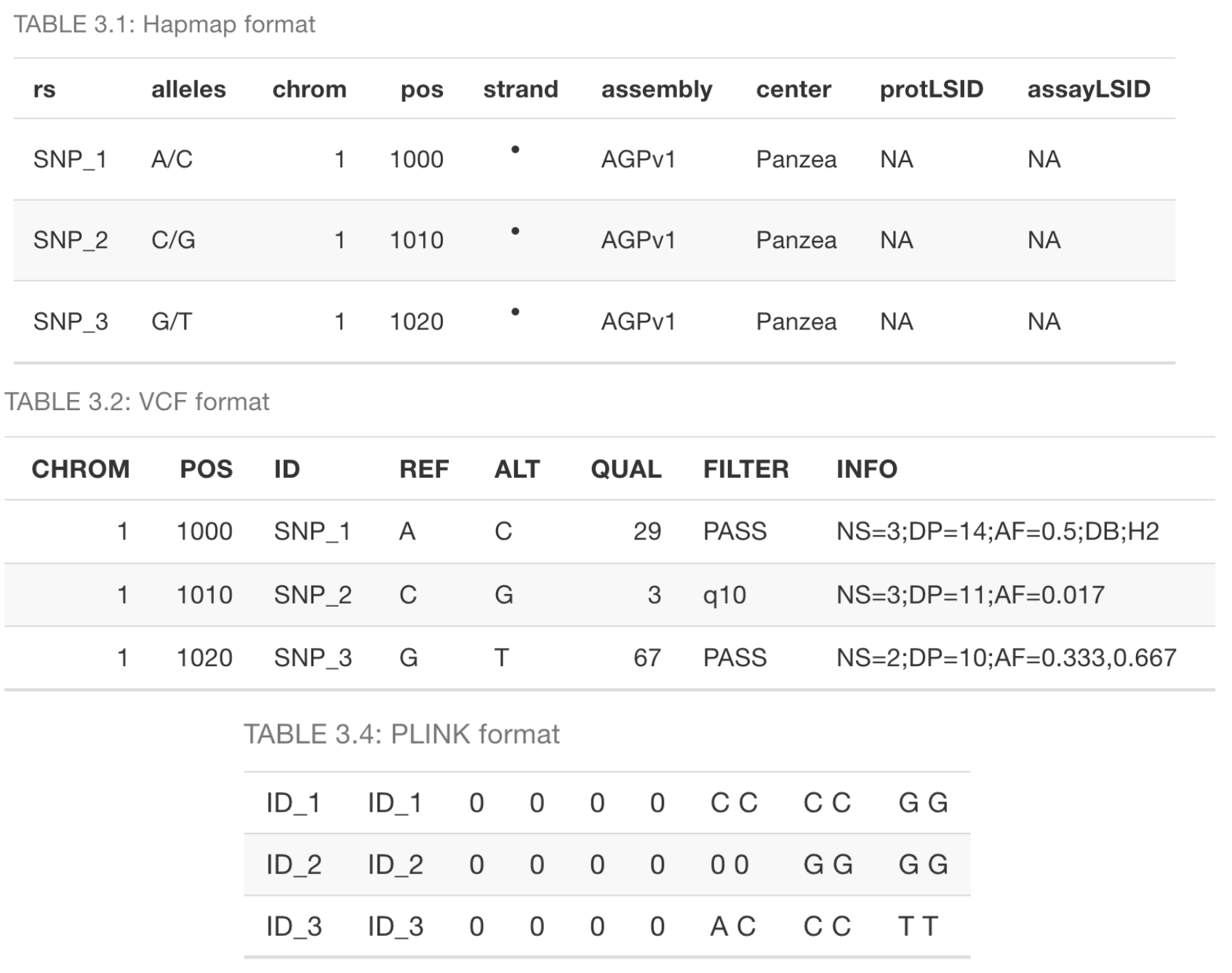
Automated Handling of Diverse Data Formats
Addressing the challenge of inconsistent format requirements across different software packages, iPat supports a wide range of genotype data formats, including Hapmap, VCF, PLINK, and numerical formats. The software automatically detects the input format based on the file content and performs necessary conversions behind the scenes, allowing users to flexibly switch between different analytical packages without the tedious process of manual data reformatting.
